Fermion#
# openfermion
import quantum_simulation_recipe as qsr
from quantum_simulation_recipe.fermion import Hydrogen_Chain
Basics#
Fermion anti-commutation relation#
Jordan-Wigner transformation: fermion and spin (qubit)#
Define raising and lowering operator \(S_j^{\pm}:=S_j^{X}\pm i S_j^Y\), then \(S_j^{+}|\downarrow\rangle=|\uparrow\rangle\), \(S_j^{+}|\uparrow\rangle=0\), \(S_j^{-}|\downarrow\rangle=0\), and \(S_j^-|\uparrow\rangle = |\downarrow\rangle\). Then, \(S_j^Z = S_j^+S_j^- - 1/2\).
They satisfy the anti-commutation relation when on the same site, \(\{S_j^+,S_j^-\}= S_j^+S_j^-+S_j^-S_j^+ = 1\).
And commute with each other for different sites, \([S_j^{\pm},S_k^{\pm}]=0\), \(j\neq k\)
Fermion operators: \(f_j^\dagger |0\rangle=|1\rangle\), \(f_j^\dagger |1\rangle=0\), \(f_j |0\rangle=0\), and \(f_j |1\rangle=|0\rangle\). Then, the number operator \(n_j=f_j^\dagger f_j\) and \(f_j^2 = (f_j^\dagger)^2 = 0\). \([n_k, n_j]=0\), \((n_k)^n=n_k\), \((n_k)^0=1\).
The anticommutation relation of fermion: not only \(\{f_j, f_j^\dagger \}=1\), but also (different site) \(\{f_j, f_k\}=\{f_j^\dagger, f_k^\dagger\} = \{f_j, f_k^\dagger\}=0\).
also written as \(\{f_j,f_k^\dagger\}=\delta_{j,k}\).
Jordan-Wigner transformation (non-local): \(S_j^- = \exp(i\pi \sum_{l=1}^{j-1} f_l^\dagger f_l) f_j\) and \(S_j^\dagger = f_j^\dagger \exp(-i\pi \sum_{l=1}^{j-1}f_l^\dagger f_l)\).
Reference:
Jordan-Wigner (Zeitschrift für Physik, 47, 631-651 (1928))
Parity (The Journal of chemical physics, 137(22), 224109 (2012))
Bravyi-Kitaev (Annals of Physics, 298(1), 210-226 (2002))
# https://qiskit-community.github.io/qiskit-nature/tutorials/06_qubit_mappers.html
from qiskit_nature.second_q.drivers import PySCFDriver
driver = PySCFDriver()
problem = driver.run()
fermionic_op = problem.hamiltonian.second_q_op()
from qiskit_nature.second_q.mappers import JordanWignerMapper
mapper = JordanWignerMapper()
print(fermionic_op)
qubit_jw_op = mapper.map(fermionic_op)
print(qubit_jw_op)
Fermionic Operator
number spin orbitals=4, number terms=36
-1.25633907300325 * ( +_0 -_0 )
+ -0.471896007281142 * ( +_1 -_1 )
+ -1.25633907300325 * ( +_2 -_2 )
+ -0.471896007281142 * ( +_3 -_3 )
+ 0.3378550774017582 * ( +_0 +_0 -_0 -_0 )
+ 0.3322908651276483 * ( +_0 +_1 -_1 -_0 )
+ 0.3378550774017582 * ( +_0 +_2 -_2 -_0 )
+ 0.3322908651276483 * ( +_0 +_3 -_3 -_0 )
+ 0.09046559989211571 * ( +_0 +_0 -_1 -_1 )
+ 0.09046559989211571 * ( +_0 +_1 -_0 -_1 )
+ 0.09046559989211571 * ( +_0 +_2 -_3 -_1 )
+ 0.09046559989211571 * ( +_0 +_3 -_2 -_1 )
+ 0.09046559989211571 * ( +_1 +_0 -_1 -_0 )
+ 0.09046559989211571 * ( +_1 +_1 -_0 -_0 )
+ 0.09046559989211571 * ( +_1 +_2 -_3 -_0 )
+ 0.09046559989211571 * ( +_1 +_3 -_2 -_0 )
+ 0.3322908651276483 * ( +_1 +_0 -_0 -_1 )
+ 0.3492868613660083 * ( +_1 +_1 -_1 -_1 )
+ 0.3322908651276483 * ( +_1 +_2 -_2 -_1 )
+ 0.3492868613660083 * ( +_1 +_3 -_3 -_1 )
+ 0.3378550774017582 * ( +_2 +_0 -_0 -_2 )
+ 0.3322908651276483 * ( +_2 +_1 -_1 -_2 )
+ 0.3378550774017582 * ( +_2 +_2 -_2 -_2 )
+ 0.3322908651276483 * ( +_2 +_3 -_3 -_2 )
+ 0.09046559989211571 * ( +_2 +_0 -_1 -_3 )
+ 0.09046559989211571 * ( +_2 +_1 -_0 -_3 )
+ 0.09046559989211571 * ( +_2 +_2 -_3 -_3 )
+ 0.09046559989211571 * ( +_2 +_3 -_2 -_3 )
+ 0.09046559989211571 * ( +_3 +_0 -_1 -_2 )
+ 0.09046559989211571 * ( +_3 +_1 -_0 -_2 )
+ 0.09046559989211571 * ( +_3 +_2 -_3 -_2 )
+ 0.09046559989211571 * ( +_3 +_3 -_2 -_2 )
+ 0.3322908651276483 * ( +_3 +_0 -_0 -_3 )
+ 0.3492868613660083 * ( +_3 +_1 -_1 -_3 )
+ 0.3322908651276483 * ( +_3 +_2 -_2 -_3 )
+ 0.3492868613660083 * ( +_3 +_3 -_3 -_3 )
SparsePauliOp(['IIII', 'IIIZ', 'IIZI', 'IZII', 'ZIII', 'IIZZ', 'IZIZ', 'ZIIZ', 'YYYY', 'XXYY', 'YYXX', 'XXXX', 'IZZI', 'ZIZI', 'ZZII'],
coeffs=[-0.81054798+0.j, 0.17218393+0.j, -0.22575349+0.j, 0.17218393+0.j,
-0.22575349+0.j, 0.12091263+0.j, 0.16892754+0.j, 0.16614543+0.j,
0.0452328 +0.j, 0.0452328 +0.j, 0.0452328 +0.j, 0.0452328 +0.j,
0.16614543+0.j, 0.17464343+0.j, 0.12091263+0.j])
Chemical molecules#
d = 1.0
n_hydrogen = 2
n = n_hydrogen * 2
h2 = Hydrogen_Chain(n_hydrogen, d, verbose=False)
# pstr: 15, ['IIII', 'ZIII', 'IZII', 'IIZI', 'IIIZ', 'ZZII', 'YXXY', 'YYXX', 'XXYY', 'XYYX', 'ZIZI', 'ZIIZ', 'IZZI', 'IZIZ', 'IIZZ']
# groups: 2
[[{'IIII': (-0.3276081896748102+0j)}, {'ZIII': (0.13716572937099497+0j)}, {'IZII': (0.13716572937099497+0j)}, {'IIZI': (-0.13036292057109025+0j)}, {'IIIZ': (-0.13036292057109025+0j)}, {'ZZII': (0.15660062488237947+0j)}], [{'YXXY': (0.049197645871367546+0j)}, {'YYXX': (-0.049197645871367546+0j)}, {'XXYY': (-0.049197645871367546+0j)}, {'XYYX': (0.049197645871367546+0j)}, {'ZIZI': (0.10622904490856078+0j)}, {'ZIIZ': (0.15542669077992832+0j)}, {'IZZI': (0.15542669077992832+0j)}, {'IZIZ': (0.10622904490856078+0j)}, {'IIZZ': (0.16326768673564335+0j)}]]
Ferimonic operators#
Pauli operators#
SYK model#
where \(\chi_i\) are majorana fermions with \(\{\chi_i,\chi_j\}=2\delta_{ij}\) and the coefficients \(J_{ijkl}\) are sampled from a Gaussian distribution \(N(0,\frac{3!}{n^3}J^2)\) with zero mean and variance..
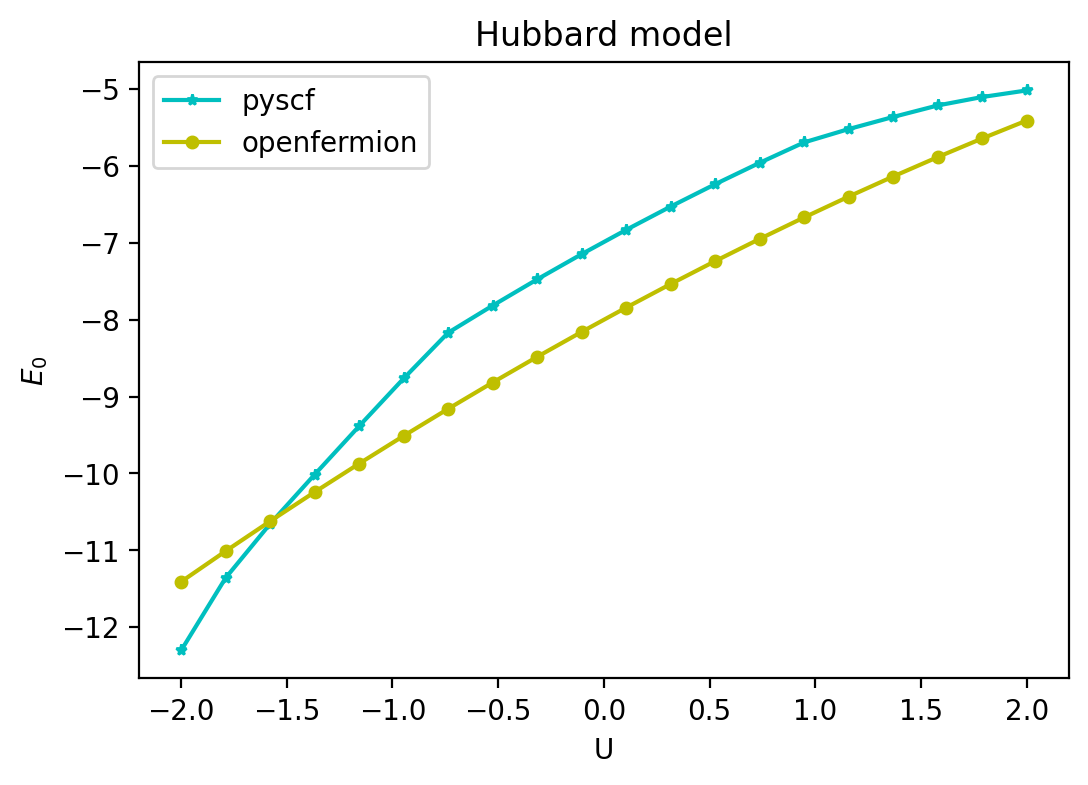
Hubbard model#
This notebook shows how to simulate the one-dimensional Fermi-Hubbard Hamiltonian
using FQE. Here \(j = 1, ..., L\) denotes site/orbital and \(\sigma \in \{ \uparrow, \downarrow \}\) denotes spin. By the end of the tutorial, we reproduce plots from the Fermi-Hubbard experiment paper and the corresponding ReCirq tutorial.
import copy
from itertools import product
import matplotlib.pyplot as plt
%config InlineBackend.figure_format = 'retina'
import numpy as np
from scipy import sparse
from scipy.linalg import expm
import openfermion as of
import fqe
from pyscf import gto, scf, ao2mo, cc, fci
"""Define the Hamiltonian."""
# Parameters.
# U = 2.0
u_list = np.linspace(-2.0, 2.0, 20)
e_list_pyscf = []
e_list_openfermion = []
def hubbard_openfermion(u):
nsites = 6
J = -1.0
hubbard = of.fermi_hubbard(1, nsites, tunneling=-J, coulomb=u, periodic=False)
sparse_hamiltonian = of.get_sparse_operator(hubbard)
ground_energy, ground_state = of.get_ground_state(sparse_hamiltonian)
# print('ground state: \n', ground_state)
return ground_energy
def hubbard_pyscf(u):
mol = gto.M(verbose=0)
n = 6
mol.nelectron = n
# Setting incore_anyway=True to ensure the customized Hamiltonian (the _eri
# attribute) to be used in the post-HF calculations. Without this parameter,
# some post-HF method (particularly in the MO integral transformation) may
# ignore the customized Hamiltonian if memory is not enough.
mol.incore_anyway = True
h1 = np.zeros((n,n))
for i in range(n-1):
h1[i,i+1] = h1[i+1,i] = -1.0
h1[n-1,0] = h1[0,n-1] = -1.0
eri = np.zeros((n,n,n,n))
for i in range(n):
eri[i,i,i,i] = u
mf = scf.RHF(mol);
mf.get_hcore = lambda *args: h1
mf.get_ovlp = lambda *args: np.eye(n)
mf._eri = ao2mo.restore(8, eri, n)
mf.kernel();
# In PySCF, the customized Hamiltonian needs to be created once in mf object.
# The Hamiltonian will be used everywhere whenever possible. Here, the model
# Hamiltonian is passed to CCSD object via the mf object.
# mycc = cc.RCCSD(mf)
# mycc.kernel()
# e,v = mycc.ipccsd(nroots=3)
# print(e)
myfci = fci.FCI(mf);
e_fci = myfci.kernel();
return e_fci[0]
for u in u_list:
e_list_pyscf.append(hubbard_openfermion(u))
e_list_openfermion.append(hubbard_pyscf(u))
# print(e_list)
fig, ax = plt.subplots(figsize=(6, 4))
ax.plot(u_list, e_list_pyscf, 'c-*', markersize=4, label='pyscf')
ax.plot(u_list, e_list_openfermion, 'y-o', markersize=4, label='openfermion')
ax.set_xlabel('U')
ax.set_ylabel(r'$E_0$')
ax.set_title('Hubbard model')
ax.legend()