Ground state#
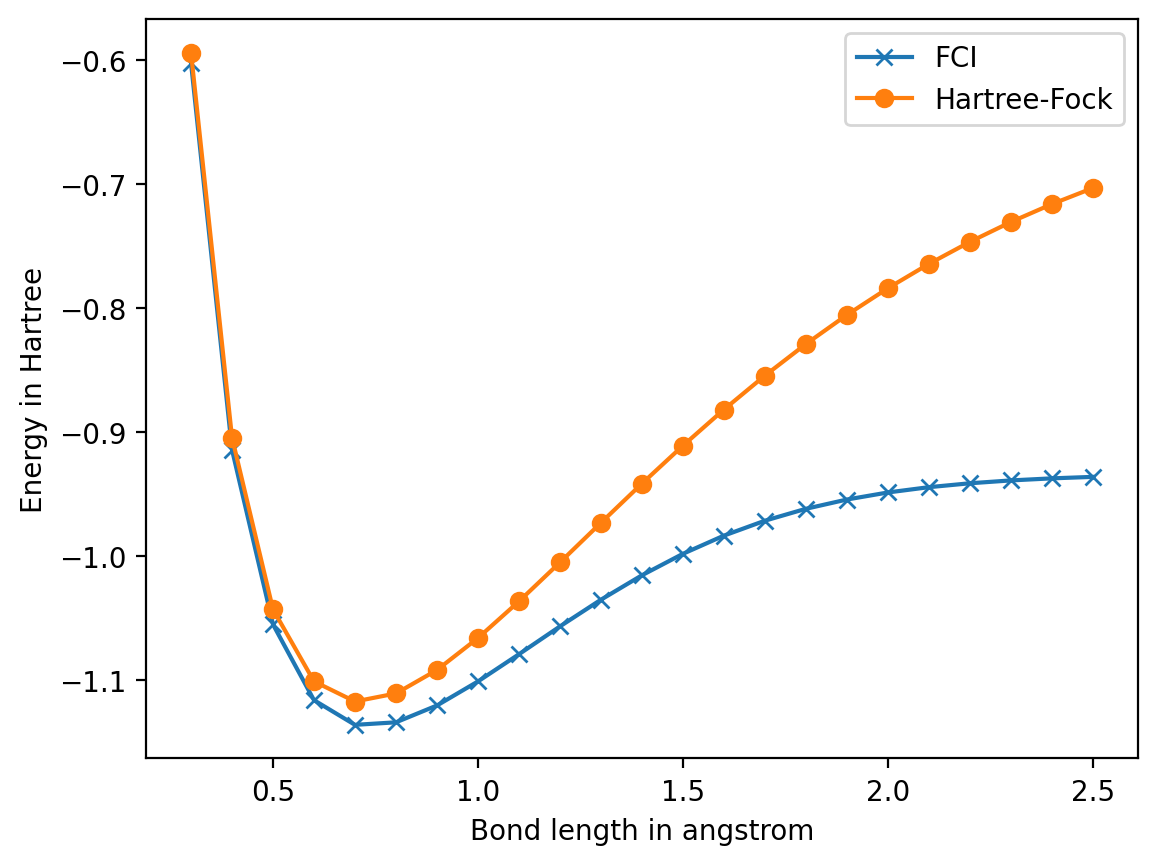
Energy surface of H chain#
https://quantumai.google/openfermion/tutorials/intro_to_openfermion
from openfermion.chem import MolecularData
# Set parameters to make a simple molecule.
diatomic_bond_length = .7414
geometry = [('H', (0., 0., 0.)), ('H', (0., 0., diatomic_bond_length))]
basis = 'sto-3g'
multiplicity = 1
charge = 0
description = str(diatomic_bond_length)
# Make molecule and print out a few interesting facts about it.
molecule = MolecularData(geometry, basis, multiplicity,
charge, description)
print('Molecule has automatically generated name {}'.format(
molecule.name))
print('Information about this molecule would be saved at:\n{}\n'.format(
molecule.filename))
print('This molecule has {} atoms and {} electrons.'.format(
molecule.n_atoms, molecule.n_electrons))
for atom, atomic_number in zip(molecule.atoms, molecule.protons):
print('Contains {} atom, which has {} protons.'.format(
atom, atomic_number))
Molecule has automatically generated name H2_sto-3g_singlet_0.7414
Information about this molecule would be saved at:
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/openfermion/testing/data/H2_sto-3g_singlet_0.7414
This molecule has 2 atoms and 2 electrons.
Contains H atom, which has 1 protons.
Contains H atom, which has 1 protons.
# Set molecule parameters.
basis = 'sto-3g'
multiplicity = 1
bond_length_interval = 0.1
n_points = 25
verbose = False
# Generate molecule at different bond lengths.
hf_energies = []
fci_energies = []
bond_lengths = []
for point in range(3, n_points + 1):
bond_length = bond_length_interval * point
bond_lengths += [bond_length]
description = str(round(bond_length,2))
geometry = [('H', (0., 0., 0.)), ('H', (0., 0., bond_length))]
molecule = MolecularData(
geometry, basis, multiplicity, description=description)
# Load data.
molecule.load()
# Print out some results of calculation.
if verbose:
print(description)
print('\nAt bond length of {} angstrom, molecular hydrogen has:'.format(
bond_length))
print('Hartree-Fock energy of {} Hartree.'.format(molecule.hf_energy))
print('MP2 energy of {} Hartree.'.format(molecule.mp2_energy))
print('FCI energy of {} Hartree.'.format(molecule.fci_energy))
print('Nuclear repulsion energy between protons is {} Hartree.'.format(
molecule.nuclear_repulsion))
for orbital in range(molecule.n_orbitals):
if verbose:
print('Spatial orbital {} has energy of {} Hartree.'.format(
orbital, molecule.orbital_energies[orbital]))
hf_energies += [molecule.hf_energy]
fci_energies += [molecule.fci_energy]
# Plot.
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(0)
plt.plot(bond_lengths, fci_energies, 'x-', label='FCI')
plt.plot(bond_lengths, hf_energies, 'o-', label='Hartree-Fock')
plt.ylabel('Energy in Hartree')
plt.xlabel('Bond length in angstrom')
plt.legend()
plt.show()